Rapid proteomic analysis for solid tumors reveals LSD1 as a drug target in an end‐stage cancer patient¶
Doll et al. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6068348/¶
Abstract¶
Recent advances in mass spectrometry (MS)‐based technologies are now set to transform translational cancer proteomics from an idea to a practice. Here, we present a robust proteomic workflow for the analysis of clinically relevant human cancer tissues that allows quantitation of thousands of tumor proteins in several hours of measuring time and a total turnaround of a few days. We applied it to a chemorefractory metastatic case of the extremely rare urachal carcinoma. Quantitative comparison of lung metastases and surrounding tissue revealed several significantly upregulated proteins, among them lysine‐specific histone demethylase 1 (LSD1/KDM1A). LSD1 is an epigenetic regulator and the target of active development efforts in oncology. Thus, clinical cancer proteomics can rapidly and efficiently identify actionable therapeutic options. While currently described for a single case study, we envision that it can be applied broadly to other patients in a similar condition.
Workflow¶

Figure 2 Proteomics workflow for the case study. (A) Timeline of the project. (B) Experimental design, including source of material, inStageTip sample preparation, and depiction of the analytical workflow
Results¶

Figure 3 Proteins differentially expressed in the urachal carcinoma lung metastases. (A) Volcano plot of the p‐values (y‐axis) vs. the log2 protein abundance differences (x‐axis) between metastases and control, with lines of significance colored in black or gray lines corresponding to a 5% or 1% FDR, respectively. (B) Mechanisms of action of LSD1/KDM1A and inhibitory drug treatment proposed: JATROSOME. TRANYLCYPROMIN
Workflow with the Clinical Knowledge Graph¶
Generate Analysis Report: Proteomics data
Identify Candidate Drug Treatments
Rank Candidates According to Toxicity
Generate Analysis Report: Proteomics Data¶
Report Manager¶
[206]:
from ckg.report_manager import project
from plotly.offline import init_notebook_mode, iplot
%matplotlib inline
init_notebook_mode(connected=True)
[207]:
configuration_files = {"proteomics":"../assets/proteomics_CS.yml"}
[208]:
study_case_project = project.Project(identifier="P0000002", configuration_files=configuration_files, datasets={}, knowledge=None, report={})
[209]:
study_case_project.build_project(force=False)
[210]:
study_case_project.generate_report()
[211]:
study_case_project.show_report(environment='notebook')
[211]:
defaultdict(list,
{'PROJECT INFORMATION': [],
'PROTEOMICS': [],
'MULTIOMICS': [],
'KNOWLEDGE GRAPH': []})
[212]:
study_case_project.list_datasets()
[212]:
dict_keys(['multiomics', 'proteomics'])
[213]:
proteomics_dataset = study_case_project.get_dataset(dataset='proteomics')
[214]:
proteomics_dataset.list_dataframes()
[214]:
['go annotation',
'number of modified proteins',
'number of peptides',
'number of proteins',
'original',
'pathway annotation',
'processed',
'protein biomarkers',
'regulated',
'regulation table',
'tissue qcmarkers']
In this case, we use the regulation table to extract proteins upregulated in the metastatic tissue compare to non-cancerous tissue.
[215]:
regulation_table = proteomics_dataset.get_dataframe(dataset_name='regulation table')
[216]:
regulation_table.head()
[216]:
| -log10 pvalue | FC | Method | T-statistics | correction | dfk | dfn | effsize | efftype | group1 | ... | identifier | log2FC | mean(group1) | mean(group2) | padj | pvalue | rejected | s0 | std(group1) | std(group2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.487413 | -1.277214 | SAMR Two class paired | 0.153337 | permutation FDR (4.0 perm) | 1 | 2 | -0.738 | hedges | CANCER | ... | A1BG~P04217 | -0.353 | 31.710 | 32.063 | 0.714951 | 0.325527 | False | 2 | 0.047 | 0.384 |
| 1 | 1.438878 | 3.610003 | SAMR Two class paired | -0.924233 | permutation FDR (4.0 perm) | 1 | 2 | 2.913 | hedges | CANCER | ... | A1CF~Q9NQ94 | 1.852 | 26.563 | 24.711 | 0.052925 | 0.036402 | False | 2 | 0.360 | 0.366 |
| 2 | 3.001177 | -1.926524 | SAMR Two class paired | 0.463202 | permutation FDR (4.0 perm) | 1 | 2 | -18.081 | hedges | CANCER | ... | A2M~P01023 | -0.946 | 34.287 | 35.233 | 0.271411 | 0.000997 | False | 2 | 0.033 | 0.026 |
| 3 | 1.192262 | 2.801113 | SAMR Two class paired | -0.594485 | permutation FDR (4.0 perm) | 1 | 2 | 2.145 | hedges | CANCER | ... | AAAS~Q9NRG9 | 1.486 | 26.317 | 24.831 | 0.149537 | 0.064230 | False | 2 | 0.173 | 0.532 |
| 4 | 0.321607 | -1.786332 | SAMR Two class paired | 0.346611 | permutation FDR (4.0 perm) | 1 | 2 | -0.496 | hedges | CANCER | ... | AACS~Q86V21 | -0.837 | 26.350 | 27.187 | 0.423692 | 0.476862 | False | 2 | 0.625 | 1.211 |
5 rows × 21 columns
[217]:
regulation_table[regulation_table['identifier']=='HDAC1~Q13547']
[217]:
| -log10 pvalue | FC | Method | T-statistics | correction | dfk | dfn | effsize | efftype | group1 | ... | identifier | log2FC | mean(group1) | mean(group2) | padj | pvalue | rejected | s0 | std(group1) | std(group2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1885 | 0.782856 | 10.367528 | SAMR Two class paired | -1.008813 | permutation FDR (4.0 perm) | 1 | 2 | 1.227 | hedges | CANCER | ... | HDAC1~Q13547 | 3.374 | 30.666 | 27.292 | 0.036735 | 0.164871 | True | 2 | 0.301 | 2.201 |
1 rows × 21 columns
[218]:
up_regulated_proteins = regulation_table.loc[(regulation_table.rejected) & (regulation_table.FC > 2), ['identifier']]
[219]:
up_regulated_proteins.shape
[219]:
(199, 1)
[220]:
'HDAC1~Q13547' in up_regulated_proteins['identifier'].tolist()
[220]:
True
Graph Database Connector¶
[221]:
from ckg.graphdb_connector import query_utils, connector
[222]:
driver = connector.getGraphDatabaseConnectionConfiguration()
[223]:
queries = query_utils.read_knowledge_queries()
1) Filter for Regulated Proteins Associated to Lung Cancer:¶
[224]:
selected_queries = query_utils.find_queries_involving_nodes(queries=queries, nodes=["Protein", "Disease"], print_pretty=True)
[225]:
selected_queries.head()
[225]:
| Name | Description | involved_nodes | involved_rels | query | example | |
|---|---|---|---|---|---|---|
| id | ||||||
| Disease | associated diseases in at least two of the pro... | get relationships to diseases from a list of p... | Protein,Disease | ASSOCIATED_WITH | MATCH (project:Project)-[:STUDIES_DISEASE]-(d:... | |
| association_disease_score | specific disease | Return the list of proteins associated to a sp... | Protein,Disease | ASSOCIATED_WITH | MATCH (protein:Protein)-[r]-(disease:Disease) ... | proteins = ['A1BG~P04217','A2M~P01023','ACACB~... |
[226]:
disease_query = selected_queries.loc["association_disease_score", "query"]
proteins = ['"{}"'.format(p) for p in up_regulated_proteins["identifier"].tolist()]
diseases = ['DOID:1324']
diseases = ['"{}"'.format(d) for d in diseases]
disease_query = disease_query.format(",".join(proteins),",".join(diseases), 1)
[227]:
proteins_associated_lung_cancer = connector.getCursorData(driver=driver, query=disease_query, parameters={})
[228]:
proteins_associated_lung_cancer.head()
[228]:
| node1 | node2 | source | type | weight | |
|---|---|---|---|---|---|
| 0 | SLC44A1~Q8WWI5 | lung cancer | DISEASES | ASSOCIATED_WITH | 1.026 |
| 1 | IRF6~O14896 | lung cancer | DISEASES | ASSOCIATED_WITH | 1.192 |
| 2 | SPINT2~O43291 | lung cancer | DISEASES | ASSOCIATED_WITH | 1.216 |
| 3 | KDM1A~O60341 | lung cancer | DISEASES | ASSOCIATED_WITH | 2.405 |
| 4 | CEACAM5~P06731 | lung cancer | DISEASES | ASSOCIATED_WITH | 3.122 |
[229]:
proteins_associated_lung_cancer.shape
[229]:
(69, 5)
2) Identify Inhibitory Drugs for those Proteins¶
[230]:
selected_queries = query_utils.find_queries_involving_nodes(queries=queries, nodes=["Protein", "Drug"], print_pretty=True)
[231]:
selected_queries.head()
[231]:
| Name | Description | involved_nodes | involved_rels | query | example | |
|---|---|---|---|---|---|---|
| id | ||||||
| Drug | associated drugs in at least two of the protei... | get relationships to drugs. Limit the result t... | Protein,Drug | ACTS_ON | MATCH (protein:Protein)-[r:ACTS_ON]-(drug:Drug... | |
| association_drug_intervention_proteins | drug intervention- protein association | Return associations between a list of proteins... | Project,Protein,Clinical_variable,Drug | HAD_INTERVENTION,ACTS_ON | MATCH (project:Project)-[]-()-[:HAD_INTERVENTI... | project_id = 'P0000002'\nproteins = ['A1BG~P04... |
| association_drug_interaction_score | drug interaction association | Return the list of drugs associated to the lis... | Protein,Drug | ACTS_ON | MATCH (protein:Protein)-[r]-(drug:Drug) WHERE ... | proteins = ['A1BG~P04217','A2M~P01023','ACACB~... |
[232]:
proteins = ['"{}"'.format(p) for p in proteins_associated_lung_cancer['node1'].tolist()]
drug_query = selected_queries.loc["association_drug_interaction_score", "query"].format(",".join(proteins), 'inhibition', 0.8)
[233]:
drugs_proposed = connector.getCursorData(driver=driver, query=drug_query, parameters={})
[234]:
drugs_proposed.head()
[234]:
| Drug_desc | action | drug_id | node1 | node2 | source | type | weight | |
|---|---|---|---|---|---|---|---|---|
| 0 | None | inhibition | DB04808 | ANG~P03950 | Neamine | STITCH | ACTS_ON | 0.800 |
| 1 | None | inhibition | DB02198 | ANG~P03950 | 2-Bromoacetyl Group | STITCH | ACTS_ON | 0.800 |
| 2 | Losartan is an angiotensin II receptor blocker... | inhibition | DB00678 | ANG~P03950 | Losartan | STITCH | ACTS_ON | 0.957 |
| 3 | Tamoxifen is a non-steroidal antiestrogen used... | inhibition | DB00675 | ANG~P03950 | Tamoxifen | STITCH | ACTS_ON | 0.990 |
| 4 | A macrolide compound obtained from Streptomyce... | inhibition | DB00877 | ANG~P03950 | Sirolimus | STITCH | ACTS_ON | 0.800 |
[235]:
drugs_proposed.shape
[235]:
(67, 8)
We can already see that CKG found the same inhibitory drug that was identified in the study case published. However, many other options are proposed and could be further ranked using other criteria.¶
[236]:
from ckg.analytics_core import utils
from ckg.analytics_core.viz import viz
[237]:
net = viz.get_network(data=drugs_proposed, identifier="inhibition_drugs", args={"source":"node1", "target":"node2", "values":"weight", "node_size":"degree","title":"Proposed drugs", "color_weight":False})
[238]:
viz.visualize_notebook_network(net["notebook"], notebook_type='jupyter', layout={'width':'100%', 'height':'700px'})
[239]:
utils.json_network_to_gml(net['net_json'], path='drug_network.gml')
[240]:
proteins = ['"{}"'.format(p) for p in proteins_associated_lung_cancer['node1'].tolist()]
drug_query = queries["association_drug_interaction_score"]["query"].format(",".join(proteins), 'inhibition', 0.9)
[241]:
drugs_proposed = connector.getCursorData(driver=driver, query=drug_query, parameters={})
[242]:
drugs_proposed.head()
[242]:
| Drug_desc | action | drug_id | node1 | node2 | source | type | weight | |
|---|---|---|---|---|---|---|---|---|
| 0 | Losartan is an angiotensin II receptor blocker... | inhibition | DB00678 | ANG~P03950 | Losartan | STITCH | ACTS_ON | 0.957 |
| 1 | Tamoxifen is a non-steroidal antiestrogen used... | inhibition | DB00675 | ANG~P03950 | Tamoxifen | STITCH | ACTS_ON | 0.990 |
| 2 | Paclitaxel is a chemotherapeutic agent markete... | inhibition | DB01229 | CDH1~P12830 | Paclitaxel | STITCH | ACTS_ON | 0.957 |
| 3 | A major primary bile acid produced in the live... | inhibition | DB02659 | CDH1~P12830 | Cholic Acid | STITCH | ACTS_ON | 0.957 |
| 4 | Calcitriol is an active metabolite of vitamin ... | inhibition | DB00136 | CDH17~Q12864 | Calcitriol | STITCH | ACTS_ON | 0.957 |
[243]:
drugs_proposed.shape
[243]:
(15, 8)
[244]:
net = viz.get_network(data=drugs_proposed, identifier="inhibition_drugs", args={"source":"node1", "target":"node2", "values":"weight", "node_size":"degree","title":"Proposed drugs", "color_weight":False})
[245]:
viz.visualize_notebook_network(net["notebook"], notebook_type='jupyter', layout={'width':'100%', 'height':'700px'})
[246]:
utils.json_network_to_gml(net['net_json'], path='drug_network_reduced.gml')
3) Identify Proposed Drug’s Known Side Effects¶
[247]:
selected_queries = query_utils.find_queries_involving_nodes(queries=queries, nodes=["Phenotype", "Drug"], print_pretty=True)
[248]:
selected_queries.head()
[248]:
| Name | Description | involved_nodes | involved_rels | query | example | |
|---|---|---|---|---|---|---|
| id | ||||||
| association_drug_sideeffects | drug side effect association | Return the list of side effects linked to drugs | Phenotype,Drug | ASSOCIATED_WITH | MATCH (sideeffect:Phenotype)-[r]-(drug:Drug) W... | drugs = ['DB00439', 'DB06196']\ndrug_side_effe... |
[249]:
drugs = drugs_proposed["drug_id"].unique()
drugs = ['"{}"'.format(d) for d in drugs]
sideeffects_query = selected_queries.loc["association_drug_sideeffects", "query"].format(",".join(drugs))
[250]:
side_effects = connector.getCursorData(driver=driver, query=sideeffects_query, parameters={})
[251]:
side_effects.head()
[251]:
| node1 | node2 | source | type | |
|---|---|---|---|---|
| 0 | Losartan | Abnormality of fluid regulation | SIDER | HAS_SIDE_EFFECT |
| 1 | Losartan | Thrombocytopenia | SIDER | HAS_SIDE_EFFECT |
| 2 | Losartan | Nausea | SIDER | HAS_SIDE_EFFECT |
| 3 | Losartan | Palpitations | SIDER | HAS_SIDE_EFFECT |
| 4 | Losartan | Arthritis | SIDER | HAS_SIDE_EFFECT |
[252]:
side_effects.groupby('node1')['node2'].count()
[252]:
node1
Atorvastatin 152
Bleomycin 51
Calcitriol 59
Cholic Acid 7
Gemcitabine 94
Glyburide 63
Losartan 120
Paclitaxel 218
Tamoxifen 106
Tolbutamide 18
Vildagliptin 22
Vorinostat 42
Name: node2, dtype: int64
[253]:
net = viz.get_network(data=side_effects, identifier="side_effects", args={"source":"node1", "target":"node2", "node_size":"degree","title":"Proposed drugs", "color_weight":False})
[254]:
side_effects.head()
[254]:
| node1 | node2 | source | type | width | |
|---|---|---|---|---|---|
| 0 | Losartan | Abnormality of fluid regulation | SIDER | HAS_SIDE_EFFECT | 1 |
| 1 | Losartan | Thrombocytopenia | SIDER | HAS_SIDE_EFFECT | 1 |
| 2 | Losartan | Nausea | SIDER | HAS_SIDE_EFFECT | 1 |
| 3 | Losartan | Palpitations | SIDER | HAS_SIDE_EFFECT | 1 |
| 4 | Losartan | Arthritis | SIDER | HAS_SIDE_EFFECT | 1 |
[255]:
utils.json_network_to_gml(net['net_json'], path='side_effects.gml')
[256]:
no_registerd_side_effects = list(set(drugs_proposed['node2'].tolist()).difference(side_effects['node1'].tolist()))
[257]:
no_registerd_side_effects
[257]:
['Trichostatin A', 'Resveratrol', 'dATP']
4) Reduce Adverse Response¶
[258]:
treatment_regimens = pd.DataFrame(['Oxaliplatin',
'Capecitabine',
'Folinic acid',
'Fluorouracil'], columns=['treatment'])
[259]:
selected_queries = query_utils.find_queries_involving_nodes(queries=queries, nodes=["Clinical_variable", "Drug"], print_pretty=True)
[260]:
selected_queries.head()
[260]:
| Name | Description | involved_nodes | involved_rels | query | example | |
|---|---|---|---|---|---|---|
| id | ||||||
| association_drug_intervention_proteins | drug intervention- protein association | Return associations between a list of proteins... | Project,Protein,Clinical_variable,Drug | HAD_INTERVENTION,ACTS_ON | MATCH (project:Project)-[]-()-[:HAD_INTERVENTI... | project_id = 'P0000002'\nproteins = ['A1BG~P04... |
| side_effects_jaccard_similarity_intervention_proposed_drugs | similarity between side effects | Return the jaccard similarity between drugs us... | Drug,Clinical_variable | HAS_SIDE_EFFECT | MATCH (d1:Drug)-[:HAS_SIDE_EFFECT]->(phenotype... | intervention = ['Capecitabine', 'Fluorouracil'... |
[261]:
drugs_proposed
[261]:
| Drug_desc | action | drug_id | node1 | node2 | source | type | weight | |
|---|---|---|---|---|---|---|---|---|
| 0 | Losartan is an angiotensin II receptor blocker... | inhibition | DB00678 | ANG~P03950 | Losartan | STITCH | ACTS_ON | 0.957 |
| 1 | Tamoxifen is a non-steroidal antiestrogen used... | inhibition | DB00675 | ANG~P03950 | Tamoxifen | STITCH | ACTS_ON | 0.990 |
| 2 | Paclitaxel is a chemotherapeutic agent markete... | inhibition | DB01229 | CDH1~P12830 | Paclitaxel | STITCH | ACTS_ON | 0.957 |
| 3 | A major primary bile acid produced in the live... | inhibition | DB02659 | CDH1~P12830 | Cholic Acid | STITCH | ACTS_ON | 0.957 |
| 4 | Calcitriol is an active metabolite of vitamin ... | inhibition | DB00136 | CDH17~Q12864 | Calcitriol | STITCH | ACTS_ON | 0.957 |
| 5 | A complex of related glycopeptide antibiotics ... | inhibition | DB00290 | LIG3~P49916 | Bleomycin | STITCH | ACTS_ON | 0.958 |
| 6 | Tolbutamide is an oral antihyperglycemic agent... | inhibition | DB01124 | GCG~P01275 | Tolbutamide | STITCH | ACTS_ON | 0.957 |
| 7 | Vildagliptin, previously identified as LAF237,... | inhibition | DB04876 | GCG~P01275 | Vildagliptin | STITCH | ACTS_ON | 0.957 |
| 8 | Glyburide is a second generation sulfonylurea ... | inhibition | DB01016 | GCG~P01275 | Glyburide | STITCH | ACTS_ON | 0.957 |
| 9 | None | inhibition | DB04297 | HDAC1~Q13547 | Trichostatin A | STITCH | ACTS_ON | 0.938 |
| 10 | Vorinostat (rINN) or suberoylanilide hydroxami... | inhibition | DB02546 | HDAC1~Q13547 | Vorinostat | STITCH | ACTS_ON | 0.987 |
| 11 | Gemcitabine is a nucleoside analog used as che... | inhibition | DB00441 | CMPK1~P30085 | Gemcitabine | STITCH | ACTS_ON | 0.991 |
| 12 | Resveratrol (3,5,4'-trihydroxystilbene) is a p... | inhibition | DB02709 | PTGES~O14684 | Resveratrol | STITCH | ACTS_ON | 0.957 |
| 13 | None | inhibition | DB03222 | TXN~P10599 | dATP | STITCH | ACTS_ON | 0.900 |
| 14 | Atorvastatin (Lipitor®), is a lipid-lowering d... | inhibition | DB01076 | THBS1~P07996 | Atorvastatin | STITCH | ACTS_ON | 0.957 |
[262]:
treatment_list = ['"{}"'.format(t) for t in treatment_regimens['treatment'].tolist()]
proposed_list = ['"{}"'.format(t) for t in drugs_proposed["drug_id"].unique()]
q = selected_queries.loc['side_effects_jaccard_similarity_intervention_proposed_drugs', 'query'].replace("INTERVENTION",",".join(treatment_list)).replace("DRUG",",".join(proposed_list))
similarity = connector.getCursorData(driver=driver, query=q, parameters={})
[263]:
q
[263]:
'MATCH (d1:Drug)-[:HAS_SIDE_EFFECT]->(phenotype1) WHERE d1.name in ["Oxaliplatin","Capecitabine","Folinic acid","Fluorouracil"] WITH d1, collect(id(phenotype1)) as treatmentData MATCH (d2:Drug)-[:HAS_SIDE_EFFECT]->(phenotype2) WHERE d2.id IN ["DB00678","DB00675","DB01229","DB02659","DB00136","DB00290","DB01124","DB04876","DB01016","DB04297","DB02546","DB00441","DB02709","DB03222","DB01076"] AND d1 <> d2 WITH d1, treatmentData, d2, collect(id(phenotype2)) as proposedData, size(apoc.coll.intersection(treatmentData, collect(DISTINCT id(phenotype2)))) AS intersection RETURN d1.name AS from, d2.name AS to, intersection, gds.alpha.similarity.jaccard(proposedData, treatmentData) AS similarity ORDER BY similarity DESC\n'
[264]:
similarity = similarity[(similarity['from'].isin(treatment_regimens['treatment'].tolist())) & (similarity['to'].isin(side_effects['node1'].unique().tolist()))]
similarity = similarity.groupby('to').mean().sort_values(by='similarity')
[265]:
similarity
[265]:
| intersection | similarity | |
|---|---|---|
| to | ||
| Cholic Acid | 5.666667 | 0.041012 |
| Tolbutamide | 11.000000 | 0.083821 |
| Vildagliptin | 15.000000 | 0.098243 |
| Calcitriol | 25.333333 | 0.156174 |
| Vorinostat | 26.000000 | 0.162747 |
| Bleomycin | 28.666667 | 0.192705 |
| Glyburide | 36.000000 | 0.219190 |
| Tamoxifen | 44.000000 | 0.223948 |
| Gemcitabine | 47.000000 | 0.256839 |
| Atorvastatin | 61.000000 | 0.277987 |
| Losartan | 61.000000 | 0.310073 |
| Paclitaxel | 97.000000 | 0.378702 |
[266]:
similarity = similarity[similarity['similarity']<0.2]
[267]:
similarity
[267]:
| intersection | similarity | |
|---|---|---|
| to | ||
| Cholic Acid | 5.666667 | 0.041012 |
| Tolbutamide | 11.000000 | 0.083821 |
| Vildagliptin | 15.000000 | 0.098243 |
| Calcitriol | 25.333333 | 0.156174 |
| Vorinostat | 26.000000 | 0.162747 |
| Bleomycin | 28.666667 | 0.192705 |
5) Check in Literature Known Links Between Disease and Drugs¶
[268]:
selected_queries = query_utils.find_queries_involving_nodes(queries=queries, nodes=["Disease", "Drug"], print_pretty=True)
[269]:
selected_queries.head()
[269]:
| Name | Description | involved_nodes | involved_rels | query | example | |
|---|---|---|---|---|---|---|
| id | ||||||
| association_drug_disease_publication | drug co-mentioned with disease in publication | Return the list of publications co-mentioning ... | Drug,Disease,Publication | MENTIONED_IN_PUBLICATION | MATCH (drug:Drug)-[r:MENTIONED_IN_PUBLICATION]... | drugs = ['DB00439', 'DB06196']\ndisease = ['D... |
| association_combination_drug_disease_publication | combinations of drugs co-mentioned with disease | Return the list of publications co-mentioning ... | Drug,Disease,Publication | MENTIONED_IN_PUBLICATION | MATCH (drug:Drug)-[r:MENTIONED_IN_PUBLICATION]... | drugs = ['DB00439', 'DB06196']\ndisease = ['D... |
[270]:
reduced_drug_list = drugs_proposed.loc[drugs_proposed["node2"].isin(similarity.index.tolist() + no_registerd_side_effects), "drug_id"]
reduced_drug_list = ['"{}"'.format(d) for d in reduced_drug_list]
drug_disease_query = selected_queries.loc['association_drug_disease_publication', 'query'].format(",".join(reduced_drug_list),",".join(diseases))
[271]:
drugs_publications = connector.getCursorData(driver=driver, query=drug_disease_query, parameters={})
[272]:
drugs_publications.head()
[272]:
| Year | linkout | node1 | node2 | type | |
|---|---|---|---|---|---|
| 0 | 1988 | https://www.ncbi.nlm.nih.gov/pubmed/2454581 | dATP | PMID:2454581 | MENTIONED_IN_PUBLICATION |
| 1 | 1992 | https://www.ncbi.nlm.nih.gov/pubmed/1562461 | Calcitriol | PMID:1562461 | MENTIONED_IN_PUBLICATION |
| 2 | 1981 | https://www.ncbi.nlm.nih.gov/pubmed/7236488 | Bleomycin | PMID:7236488 | MENTIONED_IN_PUBLICATION |
| 3 | 2007 | https://www.ncbi.nlm.nih.gov/pubmed/17583686 | Resveratrol | PMID:17583686 | MENTIONED_IN_PUBLICATION |
| 4 | 2009 | https://www.ncbi.nlm.nih.gov/pubmed/19760127 | Trichostatin A | PMID:19760127 | MENTIONED_IN_PUBLICATION |
[273]:
net = viz.get_network(data=drugs_publications, identifier="drugs_publications", args={"source":"node1", "target":"node2", "node_size":"degree","title":"Proposed drugs", "color_weight":False})
[274]:
utils.json_network_to_gml(net['net_json'], path='drugs_publications.gml')
[275]:
drugs_publications.groupby('node1')["node2"].count().plot.bar();
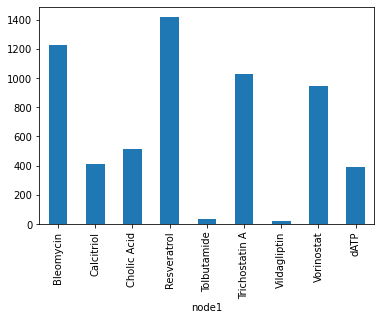
[276]:
drug_combinations_query = selected_queries.loc['association_combination_drug_disease_publication', 'query'].format(",".join(reduced_drug_list),",".join(reduced_drug_list),",".join(reduced_drug_list))
[277]:
selected_queries.loc['association_combination_drug_disease_publication', 'query']
[277]:
'MATCH (drug:Drug)-[r:MENTIONED_IN_PUBLICATION]-(publication:Publication) WHERE (drug.id IN [{}]) WITH publication, count(r) AS r_count WHERE r_count>1 MATCH (drug:Drug)-[r:MENTIONED_IN_PUBLICATION]-(publication)-[:MENTIONED_IN_PUBLICATION]-(drug2:Drug) WHERE (drug.id IN [{}]) AND drug2.id IN [{}] AND drug.id<>drug2.id RETURN (drug.name +", "+drug2.name) AS node1, publication.id AS node2, publication.linkout AS linkout, publication.year as Year, type(r) AS type\n'
[278]:
drugs_combinations = connector.getCursorData(driver=driver, query=drug_combinations_query, parameters={})
[279]:
drugs_combinations.head()
[279]:
| Year | linkout | node1 | node2 | type | |
|---|---|---|---|---|---|
| 0 | 2010 | https://www.ncbi.nlm.nih.gov/pubmed/20371703 | Cholic Acid, Calcitriol | 20371703 | MENTIONED_IN_PUBLICATION |
| 1 | 2010 | https://www.ncbi.nlm.nih.gov/pubmed/20371703 | Calcitriol, Cholic Acid | 20371703 | MENTIONED_IN_PUBLICATION |
| 2 | 2019 | https://www.ncbi.nlm.nih.gov/pubmed/31661763 | Cholic Acid, Resveratrol | 31661763 | MENTIONED_IN_PUBLICATION |
| 3 | 2019 | https://www.ncbi.nlm.nih.gov/pubmed/31661763 | Resveratrol, Cholic Acid | 31661763 | MENTIONED_IN_PUBLICATION |
| 4 | 2016 | https://www.ncbi.nlm.nih.gov/pubmed/26903812 | Cholic Acid, Calcitriol | 26903812 | MENTIONED_IN_PUBLICATION |
[280]:
drugs_combinations.groupby('node1')["node2"].count().plot.bar(figsize=(15,3));
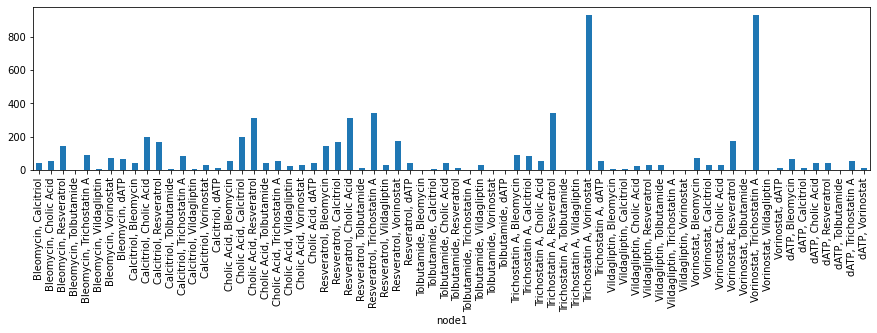
Trichostatin A (https://www.drugbank.ca/drugs/DB04297)¶
[281]:
import pandas as pd
drug_result = drugs_proposed[drugs_proposed['node2'].isin(['Trichostatin A','Vorinostat'])]
drug_result.columns = ["Drug_desc","Action","Drug_id","Protein","Drug_name","Drug_protein_source","Drug_interaction_type","Inhibition_score"]
drug_result['Side_effects'] = ";".join(side_effects[side_effects['node1']==drug_result["Drug_name"].tolist().pop()]['node2'].tolist())
drug_result['Publications'] = ";".join(drugs_publications[drugs_publications['node1']==drug_result['Drug_name'].tolist().pop()]['node2'].tolist())
protein_result = proteins_associated_lung_cancer[proteins_associated_lung_cancer['node1']==drug_result["Protein"].tolist().pop()]
protein_result.columns = ["Protein", "Disease", "Protein_disease_source", "Protein_disease_association_type", "Disease_score"]
result = pd.merge(drug_result, protein_result, on='Protein')
result = result[["Protein", "Disease",
"Protein_disease_source", "Protein_disease_association_type",
"Disease_score", "Drug_name", "Drug_id", "Drug_desc", "Action", "Drug_protein_source","Drug_interaction_type","Inhibition_score",
"Side_effects", "Publications"
]]
[282]:
result
[282]:
| Protein | Disease | Protein_disease_source | Protein_disease_association_type | Disease_score | Drug_name | Drug_id | Drug_desc | Action | Drug_protein_source | Drug_interaction_type | Inhibition_score | Side_effects | Publications | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | HDAC1~Q13547 | lung cancer | DISEASES | ASSOCIATED_WITH | 2.51 | Trichostatin A | DB04297 | None | inhibition | STITCH | ACTS_ON | 0.938 | Alopecia of scalp;Erythroderma;Syncope;Diarrhe... | PMID:32904337;PMID:31772153;PMID:27743148;PMID... |
| 1 | HDAC1~Q13547 | lung cancer | DISEASES | ASSOCIATED_WITH | 2.51 | Vorinostat | DB02546 | Vorinostat (rINN) or suberoylanilide hydroxami... | inhibition | STITCH | ACTS_ON | 0.987 | Alopecia of scalp;Erythroderma;Syncope;Diarrhe... | PMID:32904337;PMID:31772153;PMID:27743148;PMID... |
[283]:
regulation_result = regulation_table[regulation_table["identifier"]==drug_result["Protein"].tolist().pop()]
regulation_result
[283]:
| -log10 pvalue | FC | Method | T-statistics | correction | dfk | dfn | effsize | efftype | group1 | ... | identifier | log2FC | mean(group1) | mean(group2) | padj | pvalue | rejected | s0 | std(group1) | std(group2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1885 | 0.782856 | 10.367528 | SAMR Two class paired | -1.008813 | permutation FDR (4.0 perm) | 1 | 2 | 1.227 | hedges | CANCER | ... | HDAC1~Q13547 | 3.374 | 30.666 | 27.292 | 0.036735 | 0.164871 | True | 2 | 0.301 | 2.201 |
1 rows × 21 columns